RNA Interference and RNA Silencing
RNA interference (RNAi) is the process of mRNA degradation that is induced by double-stranded RNA in a sequence-specific manner. RNAi has been observed in all eukaryotes, from yeast to mammals. The RNAi pathway is thought to be an ancient mechanism for protecting the host and its genome against viruses and rogue genetic elements that use double-stranded RNA (dsRNA) in their life cycles. RNAi is now recognized to be but one of a larger set of sequence-specific cellular responses to RNA, collectively called RNA silencing. These responses have been shown to play a role not only in mRNA and dsRNA stability/degradation, but also in regulation of translation, transcription, chromatin structure, and genome integrity.
In all RNA silencing pathways, dsRNA is processed to a 21–30 nucleotide-long RNA, which then functions as a component of a “silencing complex” to specifically repress expression or function of a target gene or genomic region. One strand of the siRNA (the guide strand) is then assembled into an RNA-induced silencing complex (RISC) that cleaves the target mRNA. The siRNA is thought to provide target specificity to RISC through base pairing of the guide strand with the target mRNA.
DISCOVERY OF RNAi
RNA interference was first observed in petunias, when Napoli et al. discovered that introduction of a pigment-producing gene under control of a powerful promoter suppressed expression of both the introduced gene and the homologous endogenous gene, a phenomenon they called “cosuppression.” Cosuppression was subsequently found to occur in many species of plants and fungi (where it was called “quelling”) and to occur at the post-transcriptional level.
Short (~25 nt), antisense RNAs were first implicated in PTGS in plants. Further work in Drosophila-derived in vitro systems showed that long dsRNA was processed to short interfering RNAs (siRNAs), 21–23 nt long, that could mediate target mRNA cleavage in the region corresponding to the introduced siRNA.
Efforts to use RNAi in mammalian cells were hampered at first because of a nonspecific, interferon-mediated response to dsRNA longer than 30 bp that is seen in most mammalian cell lines. With the demonstration that 21–22 nt siRNAs, either chemically synthesized or expressed from a plasmid vector, could efficiently induce RNAi in mammalian cells without inducing the interferon response, the door was opened for development of RNAi tools in mammalian systems.
BIOGENSIS OF miRNAs AND ENDOGENOUS siRNAs
miRNAs are synthesized in the nucleus as long (up to 1000 nt) RNA polymerase II transcripts, called pri-miRNAs, that are characterized by imperfect hairpin structures. Many pri-miRNAs arise from intergenic regions, or are in antisense orientation to known genes, indicating independent transcription units. Other genes for miRNAs are found in intronic regions and could be transcribed as part of the primary transcript for the corresponding gene [42]. A dsRNA-specific endonuclease, Drosha, in conjunction with a dsRNA-binding protein, called Pasha in Drosophila and DGCR8 in humans, processes the pri-miRNA into hairpin RNAs 70–100 nt in length, called pre-miRNAs. Pre-miRNAs are transported to the cytoplasm via an Exportin-5 dependent mechanism. There Dicer works with a dsRNA-binding partner, Loqs in Drosophila and TRBP in humans, to process the pre-miRNA into mature, single-stranded miRNA and load it into an RNA silencing complex that is similar to RISC. Loqs and TRBP are functionally analogous to the dsRNA-binding protein R2D2 which partners with Dicer to process and assemble siRNAs into RISC.
THE MECHANISM OF RNA INTERFERENCE (RNAi)
 General Steps and Methods of RNA Silencing. In all RNA silencing pathways, double-stranded RNA (dsRNA) is processed to a small RNA which is assembled with RISC into a silencing complex that specifically represses expression or function of a target gene or genomic region by cleaving the corresponding mRNA.
General Steps and Methods of RNA Silencing. In all RNA silencing pathways, double-stranded RNA (dsRNA) is processed to a small RNA which is assembled with RISC into a silencing complex that specifically represses expression or function of a target gene or genomic region by cleaving the corresponding mRNA.Long double-stranded RNAs (dsRNAs; typically >200 nt) can be used to silence the expression of target genes in a variety of organisms and cell types (e.g., worms, fruit flies, and plants). Upon introduction, the long dsRNAs enter a cellular pathway that is commonly referred to as the RNA interference (RNAi) pathway. First, the dsRNAs get processed into 20-25 nucleotide (nt) small interfering RNAs (siRNAs) by an RNase III-like enzyme called Dicer (initiation step). Then, the siRNAs assemble into endoribonuclease-containing complexes known as RNA-induced silencing complexes (RISCs), unwinding in the process. The siRNA strands subsequently guide the RISCs to complementary RNA molecules, where they cleave and destroy the cognate RNA (effecter step). Cleavage of cognate RNA takes place near the middle of the region bound by the siRNA strand.

In mammalian cells, introduction of long dsRNA (>30 nt) initiates a potent antiviral response, exemplified by nonspecific inhibition of protein synthesis and RNA degradation. The mammalian antiviral response can be bypassed, however, by the introduction or expression of siRNAs.
APPLICATION OF RNAi ON VIRAL DISEASE
Viral disease is one of the most threats to the human health and also to the domestic animals. Bacterial infection can be cured by antibiotics but can’t be in the case of virus. Some viral diseases like AIDS, hepatitis B virus are very lethal to the humans which may even cause death of the infected person. There is no drug developed against the HIV virus till now and hence once infected with HIV it ends with the death of the person. In order to cure such type of disease, RNAi pose a very good technique in inhibiting such viral infections. Before going to the technique of RNAi induced treatment to viral disease let’s start with the life cycle of the virus which is explained in the lower section.
Viruses
Viruses are similar to other living organisms, however there are differences. One of the ways a virus can be seen as living is that a virus needs to replicate and create progeny. However, unlike other organisms, a virus cannot live on its own. It is only active when replicating within a host, using a hosts' resources and food. Once inside a host, a virus's sole purpose is to make as many copies of it, and infect other host cells; everything it does is to benefit its fitness and increase the number of its offspring. Generally, viruses are not regarded as living organisms as they are unable to live on their own.
MODE OF ACTION OF VIRUS
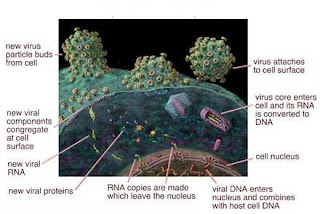 The life cycle of viruses may be divided into the following stages:
The life cycle of viruses may be divided into the following stages:1. Attachment : Attachment is a specific binding between viral surface proteins and their receptors on the host cellular surface. This specificity determines the host range of a virus. For instance, the human immunodeficiency virus (HIV) attacks only human's immune cells (mainly T cells), because its surface protein, gp120, can interact with CD4 and chemokine receptors on the T cell's surface.
2. Penetration: Following attachment, viruses may enter the host cell through receptor mediated endocytosis or other mechanisms. A virus must now enter the cell, which is covered by a phospholipid bilayer. Viral receptors attach to the receptors on the surface of the cell and secondary receptors may be present to initiate the puncture of the cell membrane or fusion with the host cell, followed by the unfolding of the viral envelope.
3. Uncoating: Uncoating is a process that viral capsid is degraded by viral enzymes or host enzymes. In essence, the virus's envelope blends with the cell membrane, releasing its contents into the cell. Obviously, this can only be done with viruses that contain an envelope. Examples include HIV, influenza.
4. Replication: As soon as the genetic material of virus enters the cytoplasm it will be carried to the nucleus where it gets integrated in to the host genome. If the genetic material is RNA then cDNA will be produced by reverse transcriptase enzyme then it is incorporated into the host genome by integrase enzyme. Now the virus will produce its own mRNA and further leads to the production of viral proteins. Also many copies of viral genome will be produced and their assembly will takes place in the cytoplasm of the host cell as shown in the fig,.
5.Release: After they get assembled,
· Viruses may escape from the host cell by causing cell rupture (lysis).
· Enveloped viruses (e.g., HIV) typically "bud" from the host cell. During the budding process, a virus acquires the phospholipid envelope containing the embedded viral glycoprotein.
The above process discussed will be recycled in order to produce more number of viruses, which is shown diagrammatically below.
From the above described life cycle of the virus we came to know about how the virus replicates inside the host cell. What ever may be the mode of infection of the virus ultimately it will be producing the mRNA, which in turn will produce the protein. The RNAi technique explains how to block or salience a specific gene using siRNA or dsRNA. If the above RNAi technique is applied on the viral disease then the deadly disease like AIDS, hepatitis B which will pose a great challenge in the medical field can be cured very easily. Although the process seems to be easy but there are many obstacles occur in transferring the siRNA or dsRNA in to the body which we will study in detail in the later session.
SUPPRESSION OF VIRAL mRNA BY RNAi TECHNIQUE



Mechanism of RNAi to degrade viral mRNA. As viral genome enters the host nucleus it produces mRNA. These are sent to cytoplasm for translation. The double stranded (ds) RNA will supplied which will be having same nucleotides as in the mRNA, into the cell The cellular enzyme dicer binds to the dsRNA and cuts it into short pieces of 20 or so nucleotide pairs in length known as small interfering RNAs or siRNAs. These bind to a cellular enzyme complex RISC (RNA induced silencing complex) that uses one strand of the siRNA to bind to single stranded RNA molecules such as mRNA of complementary sequence. RISC then degrades the mRNA, thus silencing expression of the viral gene. In mammals, other antiviral responses to dsRNA also exist.
To cure the viral disease, the RNAi technique can be used as explained below.
Ø The virus comes and attaches to the receptors present on the surface of the host cell.
Ø Now the genetic material will be transformed into the host cell and then it gets into the nucleus of the cell.
Ø The viral genome, with the help of its own enzyme, gets integrated into the host genome as shown in fig.
Ø The viral genome starts producing the mRNA and releases it to the cytoplasm.
Ø The viral infected genome is transformed with the, the dsRNAs get processed into 20-25 nt siRNAs by an RNase III-like enzyme called Dicer.
Ø Then, the siRNAs assemble into endoribonuclease-containing complexes known as RISCs, unwinding in the process.
Ø The siRNA strands subsequently guide the RISCs to complementary RNA molecules, where they cleave and destroy the cognate RNA (effecter step).
Ø Cleavage of cognate RNA takes place near the middle of the region bound by the siRNA strand.
Ø Thus the protein coded by the mRNA will not be produced, which inhibit the synthesis of enzymes and other important proteins which helps the virus to replicate.
Ø The final outcome of the whole process is, virus will not be synthesized and thus avoiding the spread of the disease in the body.
Although the process seems to be easy the delivery of siRNA to humans has many risks.
CONSTRUCTION OF siRNA EXPRESSION VECTORS
The mammalian expression vector, for each siRNA expression plasmid, a pair of 64 nucleotide oligonucleotides were annealed to create a 5' Bgl-II and 3' Hind III site flanking a 21 nt target sequence in the de-branching enzyme (DBR1) gene, a central 9 nt loop followed by a 21 nt antisense copy of the target sequence. Three pairs of cDNA oligonucleotides, targeting the human DBR1 gene at different locations were synthesized by MWG Biotech (Irvine, CA). Each pair of oligonucleotides was annealed at 90°C for 4 minutes, then at 70°C for 10 minutes, cooled to 37°C, and incubated for 20 minutes. The annealed dsDNA oligonucleotides were ligated into the pHyper vector between the Bgl-II and Hind III sites. These three 21 nt DBR1 sequences, D1, AACGAGGCGGATCTACGCTGC; D2, AAGGATCGGTGGAATCTCTGG; D3, AATGTGACTGGGCGCCTGTGG, were used to create three DBR1-siRNA plasmids, designated pHyper-D1, D2 and D3.
DELIVERY OF siRNA IN VIVO
The Two Basic Methods
1. Local Administration of siRNAs
Local administration means restricted to specific part. Perhaps the greatest success has come with local administration of siRNA to the eye. A siRNA delivered to the subretinal space in mice has been proven to reduce eye angiogenesis. In this model, there was no need to modify the siRNA, or to complex it with lipid polymer or other delivery agents, as has been necessary in some other systems. They have also successfully delivered locally the siRNA to the rodent brain to cure a neuropathic pain. In one promising study, an intranasal delivery system was used in primates to deliver a SARS virus specific siRNA, resulting in reduced fever, decreased viral load, and reduced alveoli damage. This study demonstrates the validity of intranasal delivery of siRNAs to the lung.
2. Systemic siRNA Delivery
The work on systemic delivery of siRNAs illustrates three obstacles that must be overcome for siRNA to be successful in vivo.
1.Selective delivery into the desired tissue.
2.Adequate protection from degradation en route to the target tissue.
3.And protection of the siRNA from rapid excretion.
Surprisingly, rapid excretion has proven to be more of a problem than in vivo stability. Although chemical stabilization is readily achieved via siRNA modification, it does not appear to be necessary in most cases, as excretion appears to occur prior to degradation. Use of nanoparticles or lipid complexes currently shows more promise than chemical modification to address the pharmacokinetics and tissue distribution issues endemic to in vivo siRNA delivery.
More recently, low volume, normal pressure intravenous delivery of a modified siRNA targeting apolipoprotein B in mice resulted in gene silencing in the liver and jejunum. The siRNA was conjugated with cholesterol to provide targeted delivery, and included backbone and sugar modifications to enhance serum stability.
OVERCOMING SAFTY AND SPECIFICITY CONCERNS
Obviously as with all new therapeutic modalities, the safety of the approach is an essential parameter that requires an utmost thorough preclinical evaluation. In the case of RNAi, the earliest reports of adverse effects date back to 2003, shortly after the first demonstration of RNAi efficacy in human cells, when multiple groups found an induction of IFN responses in siRNA-treated cells. The initial enthusiasm further waned with additional findings of off-target effects from siRNAs and shRNAs, i.e., the unwanted downregulation of nontargeted mRNAs. Subsequent large-scale gene expression profiling studies confirmed that partial sequence homologies were sufficient to induce silencing of off-target genes, in many cases thought to occur through an miRNA-mediated mechanism. Adding to the concerns are recent reports of siRNA-lipid complex–induced stimulation of TLRs, especially TLR7, in plasmacytoid dendritic cells. Importantly, it was also found that this effect is cell- and sequence-specific and the result of particular offending GU-rich “danger motifs” in siRNAs, suggesting that this effect can be avoided by using optimized siRNAs and formulations that do not target these cells. Similarly, multiple reports have shown that chemical modifications of siRNAs can abrogate IFN and cytokine induction. This further adds to the hope that such side effects can be eliminated altogether in future clinical protocols. Important to note is that the majority of RNAi-associated adverse immune responses have only been observed in cultured cells thus far, and their relevance in vivo remains unclear. Moreover, based on the complexity and differences in the innate responses between animals and humans, further clinical testing of siRNA delivery complexes is required before an accurate assessment of their therapeutic index can be established.
OTHER APPLICATION OF RNAi
Macular Degeneration
It makes sense that the first RNAi therapy to reach patients in clinical trials would aim at a debilitating eye disease called macular degeneration. Biotech firms had set their sights on the disease for many reasons: Most critically, RNAi drugs can be delivered directly to the diseased tissue—literally injected into the eye. This direct delivery helps ensure that "naked" RNAi drugs—short strands of RNA that aren't packaged and protected in membranes and which quickly break down in the bloodstream—can reach their target intact. Local delivery also makes it less likely that the drugs will have unanticipated, harmful effects elsewhere in the body.
What's more, the disease is triggered by a well-known culprit—a protein called VEGF that promotes blood vessel growth. In patients with macular degeneration, too much of this protein leads to the sprouting of excess blood vessels behind the retina. The blood vessels leak, clouding and often entirely destroying vision. The new RNAi drugs shut down genes that produce VEGF and allow it to make the leaky vessels.
Cancer
Cancer often involves mutant genes that promote uncontrolled cell growth. In the last few years, researchers have silenced more than a dozen known cancer-causing genes with RNAi. Yet, once again, most of this success has been with cell cultures in the lab, and delivery poses the key hurdle in moving from the lab to the bedside of patients. Researchers are just beginning, for instance, to sort through how RNAi therapies might reach and penetrate tumors.
Rather than take a leading role, some RNAi therapies may help defeat cancers by supporting chemotherapy. Drug resistance is a major problem in chemotherapy, thwarting between 20 and 50 percent of all current treatments. In many of these failures, the guilty agent is a protein called P-glycoprotein. Like a misguided housekeeper, this protein sweeps drugs out of diseased cells. In 2004, a team of scientists at Imperial College London showed that RNAi can stop production of the protein in multidrug-resistant leukemia cells, restoring their sensitivity to existing drugs.
RNAi also provides a powerful new way for scientists to discover and learn more about genes that trigger or inhibit cancer. Greg Hannon and his group at Cold Spring Harbor Laboratory are part of an effort to decipher the function of 15,000 genes in a variety of human cancer cell lines. Such efforts might pinpoint genes never before linked to cancer and generate novel ideas for treatments.
LOOKING TO THE FUTURE
RNAi is now firmly entrenched as an invaluable tool in most drug discovery pipelines, and further advances in the technology will no doubt enhance the utility of the technique in identifying and validating potential drug targets in vivo. In addition, siRNAs themselves have huge potential as therapeutic agents. If realized, the impact on the pharmaceutical industry would be revolutionary. The most significant hurdle for the therapeutic use of siRNAs is how to provide targeted delivery. Although significant progress has been made, delivery of nucleic acids to specific organs, tissues, and cells will require additional advances, including development of possible novel conjugations and/or formulations to specifically target certain cells.
CONCLUTION
However, as the field rapidly matures, we are also beginning to realize that routine clinical mRNA targeting might not yet be entirely ready for prime time. Despite the optimism and excitement over the first results from clinical evaluations of locally applied siRNAs, it must not be overlooked that new concerns are constantly being unraveled by preclinical studies, and unknown further roadblocks might lie ahead. Therefore, in particular for challenging chronic diseases such as cancer or viral infections, therapeutic RNAi is probably still facing a bumpy road toward proving its worth in looming clinical trials. All this raises considerable hope that a revolutionary transformation in modern medicine is on the horizon and that an arsenal of effective and safe systemic RNAi therapies for a wide range of human diseases may indeed be realized within a few years.
REFERENCES
1.Downward J (2004) Use of RNA interference libraries to investigate oncogenic signalling in mammalian cells. Oncogene 23(51):8376–8383.
2. Filipowicz W (2005) RNAi: the nuts and bolts of the RISC machine. Cell 122(1):17–20.
3. Sachse C, Krausz E, Kronke A, Hannus M, Walsh A, Grabner A, Ovcharenko D, Dorris D, Trudel C, Sonnichsen B, Echeverri CJ (2005) High-throughput RNA interference strategies for target discovery and validation by using synthetic short interfering RNAs: functional genomics investigations of biological pathways. Methods Enzymol 392:242–277.
4. Sontheimer EJ, Carthew RW (2005) Silence from within: endogenous siRNAs and miRNAs. Cell 122(1):9–12.
5. Tomari Y, Zamore PD (2005) Perspective: machines for RNAi. Genes Dev 19(5):517–529.
6. Zamore PD, Haley B (2005) Ribo-gnome: the big world of small RNAs. Science 309(5740):1519–1524.
7. Sontheimer EJ (2005) Assembly and function of RNA silencing complexes. Nat Rev Mol Cell Biol 6(2):127–138.
8. Napoli C, Lemieux C, Jorgensen R (1990) Introduction of a Chimeric Chalcone Synthase Gene into Petunia Results in Reversible Co-Suppression of Homologous Genes in trans. Plant Cell 2(4):279–289.
9. Cogoni C, Irelan JT, Schumacher M, Schmidhauser TJ, Selker EU, Macino G (1996) Transgene silencing of the al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA-DNA interactions or DNA methylation. Embo J 15(12):3153–3163.
10. Cogoni C, Macino G (2000) Post-transcriptional gene silencing across kingdoms. Curr Opin Genet Dev 10(6):638–643.
11. Palauqui JC, Vaucheret H (1998) Transgenes are dispensable for the RNA degradation step of cosuppression. Proc Natl Acad Sci USA 95(16):9675–9680.
12. Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391(6669):806–811.
13. Hamilton AJ, Baulcombe DC (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286(5441):950–952.
14. Elbashir SM, Lendeckel W, Tuschl T (2001) RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev 15(2):188–200.
15. Hammond SM, Bernstein E, Beach D, Hannon GJ (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404(6775):293–296.
16. Zamore PD, Tuschl T, Sharp PA, Bartel DP (2000) RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101(1):25–33.
17. Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J (2001) Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol 2(1):2.1–2.10.
18. Tabara H, Grishok A, Mello CC (1998) RNAi in C. elegans: soaking in the genome sequence. Science 282(5388):430–431.
19. Timmons L, Court DL, Fire A (2001) Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditiselegans. Gene 263(1–2):103–112.
20. Armknecht S, Boutros M, Kiger A, Nybakken K, Mathey-Prevot B, Perrimon N (2005) High-throughput RNA interference screens in Drosophila tissue culture cells. Methods Enzymol 392:55–73.
21. Brummelkamp TR, Bernards R, Agami R (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science 296(5567):550–553.
22. Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411(6836):494–498.
23. Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75(5):843–854.
24. Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403(6772):901–906.
25. Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T (2001) Identification of novel genes coding for small expressed RNAs. Science 294(5543):853–858.
26. Lau NC, Lim LP, Weinstein EG, Bartel DP (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294(5543):858–862.
27. Lee RC, Ambros V (2001) An extensive class of small RNAs in Caenorhabditis elegans. Science 294(5543):862–864.
28. Llave C, Kasschau KD, Rector MA, Carrington JC (2002) Endogenous and silencing-associated small RNAs in plants. Plant Cell 14(7):1605–1619.
29. Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP (2002) MicroRNAs in plants. Genes Dev 16(13):1616–1626.
30. Ambros V (2004) The functions of animal microRNAs. Nature 431(7006):350–355.
31. Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM (2004) Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA 101(9):2999–3004.
32. Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ (2005) RAS is regulated by the let-7 microRNA family. Cell 120(5):635–647.
33. Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, Sharon E, Spector Y, Bentwich Z (2005) Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet 37(7):766–770.
34. Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120(1):15–20.
35. Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409(6818):363–366.
36. Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD (2001) A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293(5531):834–838.
37. Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH (2001) Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev 15(20):2654–2659.
38. Knight SW, Bass BL (2001) A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293(5538):2269–2271.
39. Aravin AA, Klenov MS, Vagin VV, Bantignies F, Cavalli G, Gvozdev VA (2004) Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. Mol Cell Biol 24(15):6742–6750.
40. Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D (2003) MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol 13(10):807–818.
41. Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crete P (2004) Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell 16(1):69–79.
42. Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN (2004) MicroRNA genes are transcribed by RNA polymerase II. Embo J 23(20):4051–4060.
43. Siolas D, Lerner C, Burchard J, Ge W, Linsley PS, Paddison PJ, Hannon GJ, Cleary MA (2005) Synthetic shRNAs as potent RNAi triggers. Nat Biotechnol 23(2):227–231.
44. Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD (2005) Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 123:607–620.
45. Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC (2005) Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev 19(23):2837–2848.
46. Leuschner PJF, Ameres SF, Kueng S, Martinez J (2006) Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep AOP 20 January 2006.
47. Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L (2005) Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol 12(4):340–349.
47a. Gregory RI, Chendrimada TP, Cooch N, and Shiekhattar R. (2005) Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell(123):631–640.
48. Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W (2005) Inhibition of translational initiation by Let-7 microRNA in human cells. Science 309(5740):1573–1576.
49. Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D (2004) RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303(5658):672–676.
50. Qi Y, Denli AM, Hannon GJ (2005) Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell 19(3):421–428.
51. Check E (2005) A crucial test. Nat Med 11(3):243–244.
52. Schmidt C. Knockout Punch: The Promise of RNAi. bio.com, New & Features 2005 Jun 28 [cited; Available online
53. Dasgupta R, Perrimon N (2004) Using RNAi to catch Drosophila genes in a web of interactions: insights into cancer research. Oncogene 23(51):8359–8365.
54. Poulin G, Nandakumar R, Ahringer J (2004) Genome-wide RNAi screens in Caenorhabditis elegans: impact on cancer research. Oncogene 23(51):8340–8345.
55. Timmons L, Fire A (1998) Specific interference by ingested dsRNA. Nature 395(6705):854.
56. Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421(6920):231–237.
57. Rual JF, Ceron J, Koreth J, Hao T, Nicot AS, Hirozane-Kishikawa T, Vandenhaute J, Orkin SH, Hill DE, van den Heuvel S, Vidal M (2004) Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res 14(10B):2162–2168.
58. Muller P, Kuttenkeuler D, Gesellchen V, Zeidler MP, Boutros M (2005) Identification of JAK/STAT signalling components by genome-wide RNA interference. Nature 436(7052):871–875.
59. Silva J, Chang K, Hannon GJ, Rivas FV (2004) RNA-interference-based functional genomics in mammalian cells: reverse genetics coming of age. Oncogene 23(51):8401–8409.
60. Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, Kerkhoven RM, Madiredjo M, Nijkamp W, Weigelt B, Agami R, Ge W, Cavet G, Linsley PS, Beijersbergen RL, Bernards R (2004) A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature 428(6981):431–437.
61. Paddison PJ, Silva JM, Conklin DS, Schlabach M, Li M, Aruleba S, Balija V, O’Shaughnessy A, Gnoj L, Scobie K, Chang K, Westbrook T, Cleary M, Sachidanandam R, McCombie WR, Elledge SJ, Hannon GJ (2004) A resource for large-scale RNA-interference-based screens in mammals. Nature 428(6981):427–431.
No comments:
Post a Comment